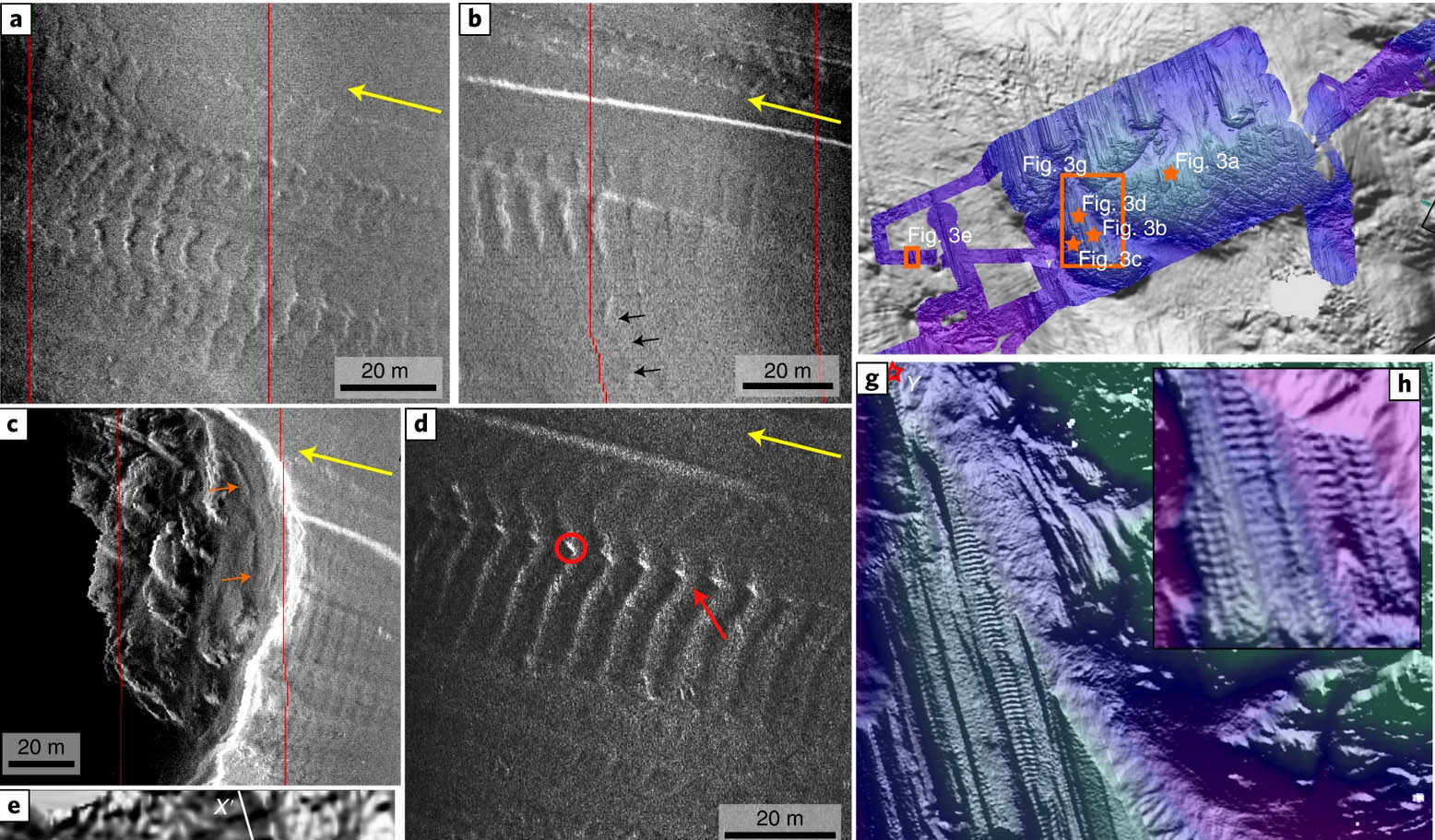
A claim today that humans evolved from worms is based on scientists discovering a proto-backbone in the world’s oldest worm fossil.
However as with all great science news headlines, it’s never as simple as those seeking funding and the news media who report the discoveries try to make it.
The assumption is made that “because” creature A has a feature that creature C has, creature A “must” have evolved into creature C. Yet biologists have long known that nature repeatedly throws up examples of similar creatures that look the same (bees and wasps, for example) but who don’t share common ancestors.
The phenomenon is known as “convergent evolution“, and is often expressed:
A -> B
C -> B
A feature “B”, such as a spine in this instance, may have developed separately in vastly different organisms (A and C). The discovery of a proto-backbone in an ancient fossil therefore does not prove an evolutionary pathway to anywhere.
The picture of sharks and dolphins shows the body form of totally unrelated species can be explained simply by adaptation, not inheritance.
Yet ‘humans evolved from worms” still makes a great headline, and you wouldn’t be reading this if it didn’t.
Meanwhile, if you are after a more significant development in the evolution debate, try this from the Discovery Institute:
The epigenome looks like it could be the evolutionists’ newest nightmare — and the latest icon of intelligent design. Back in the 1950s, the genome coded in DNA could well have finished off Darwin. Its digital code, faithfully copied and reproduced by a host of molecular machines, was not the kind of sophistication that Darwinian theory expected, or seemed capable of explaining. Nevertheless, fancy footwork and rhetorical swordsmanship has kept the theory in a standoff with versions of ID for some sixty years.
Enter the epigenome, with its codes, upon codes, upon codes. Discovery Institute’s Richard Sternberg has made this the focus of his research lately and, as Casey described the other day, it’s also the subject of a new book based on Sternberg’s work, by Tom Woodward and James Gills, The Mysterious Epigenome: What Lies Beyond DNA.
When all else fails, look brave. Three Harvard biologists, Ben Hunter, Jesse D. Hollister, and Kirsten Bomblies, have now sought to face the daunting new challenge with an essay in Current Biology, “Epigenetic Inheritance: What News for Evolution?”
The epigenome has no clear boundaries at this point. “Epigenetics” could refer to any heritable condition beyond DNA. Scientists now know that gene expression is controlled by numerous mechanisms, including histone tags (sometimes called the “histone code”), the “zygote code,” various types of small RNAs, alternative splicing, pre- and post-transcriptional modification, and an army of transcription factors. The old “central dogma” that information flows from gene to protein has been defunct for some time, but much remains to be understood. How does the environment influence epigenetic markers? How are they passed on, and how stable are they? How do “epialleles” (a new term extending Mendel’s paired gene concept) affect the phenotype?
Epigenetic marks such as cytosine methylation or histone modifications can be very dynamic and can alter gene expression in response to environmental and developmental cues without changes in DNA sequence; in some cases epigenetic changes can be heritable through meiosis. This has spurred interest — and heated debates — about whether epigenetic variation may play a significant role in adaptive evolution. The need to formally consider epialleles in population genetics and evolutionary theory has been emphasized … however, more empirical data are necessary to parameterize models and assess the actual impacts of epigenetic variation on adaptive phenotypes.
So far, only a couple of transgenerational studies on the lab plant Arabidopsis thaliana provide detailed data on long-term inheritance of epigenetic tags. They show that some tags are dynamic and some static, with the static markers tending to reside with non-coding regions of DNA. There appear to be “hotspots” for epigenetic change. Some markers can revert; others appear to be metastable. Without a clear view of the threat (just the echo of its approaching footsteps), the Harvard team decided to engage in possibility thinking. Maybe it’s something they can throw the “epigenetic junk” weapon at.
It has been known for some time that epialleles at some loci are “metastable” and can change dramatically over generations. Such instability suggests it is unlikely that alternative epialleles can contribute appreciably to stable evolutionary change.
While instability speaks against the idea that individual epialleles would contribute to long-term adaptive evolution, it does beg the question why there is variation among loci in epigenetic stability in the first place.